Unmasking Vulnerabilities in the Age of COVID-19: A Comprehensive Review
Graphical Abstract
Abstract
The COVID-19 pandemic, driven by the novel
coronavirus SARS-CoV-2, has evolved into an unprecedented global health crisis,
encompassing zoonotic origins, a shift in human-to-human transmission dynamics
and global dissemination, The review navigates through the complexities of
COVID-19. Beyond mild respiratory symptoms, the clinical spectrum includes
severe conditions like multi-organ failure, pneumonia and acute respiratory
distress syndrome (ARDS). Critical evaluation of COVID-19 diagnostic
techniques, including PCR, antigen tests and serological assays, emphasizes
their pivotal role in disease detection, management, contact tracing and
containment. In the therapeutic domain, the review explores treatments such as
antivirals, immunomodulatory therapies and repurposed pharmaceuticals, with a
spotlight on vaccine development for epidemic containment and herd immunity.
Despite progress, global healthcare systems face formidable challenges,
including equitable vaccine distribution, disinformation combat, viral mutation
management and strategic planning for future outbreaks. A comparative analysis
of SARS highlights the need to distinguish between these diseases for effective
epidemic management. The study aims to provide profound insights into the
diverse nature of COVID-19, fostering a deeper understanding and guiding future
research and public health initiatives.
Keywords: COVID-19, Zoonotic spillover, Pandemic, Emerging variants, SARS-CoV-2, Vaccine,
Clinical manifestations
1. Introduction
The
SARS-CoV-2 virus, which caused the SARS epidemic in 2002–2004, is the source of
the current pandemic, which is still lurking. Real-time observation of ongoing
evolutionary processes has provided a significant understanding of SARS-CoV-2
diversification. Numerous variations have emerged as a result of this
diversification, each set apart by unique traits like immunological evasion,
severity and transmissibility. Changes in immune profiles, human migration and
infected individuals are all part of the complex evolutionary path that is
intimately connected to ecological dynamics and the events of transmission1. Among the 180 identified species of RNA viruses capable
of infecting humans, an average of two new species emerge each year. RNA
viruses have extensively spread among humans, other mammals and occasionally
birds, across both epidemiological and evolutionary timelines. Notably, 89% of
human-infective species are zoonotic and a considerable proportion of the
remaining species trace their origins back to zoonotic sources. The pace at
which mutations are created and propagated across populations is the most
important factor in viral evolution2. Natural selection also helps to fix favourable
mutations and improve transmissibility3. However, viral evolution becomes complicated when
viruses reproduce and develop inside humans while adapting to effective human-to-human
transmission. As viral lineages evolve, antigenically distinct strains may
emerge at higher organizational levels4. This article aims to explore the evolutionary dynamics
of SARS-CoV-2 across various scales. This encompasses examining the stages of
the COVID-19 (coronavirus disease 2019) pandemic, identifying crucial factors
influencing the virus's evolution, exploring hypotheses surrounding the
emergence of statistically significant variants and contemplating potential
evolutionary pathways that could impact public health in the future.
Considering the substantial role of the SARS-CoV-2 virus in triggering the
COVID-19 pandemic, a comprehensive investigation into the infection and its
repercussions for public health is essential. The review also delves into the
transmission of the SARS-CoV-2 virus from patient to host, the utilization of
mathematical models for predicting the risk of viral aerosol/droplet
transmission, potential pathways for viral entry into the human host and the
cellular mechanisms underlying these processes.
Also,
highlights COVID-19 clinical symptoms and available diagnostic approaches for
detecting the virus. The requirement for effective treatment techniques, such
as vaccine development and medication repurposing, is emphasized here. Given
the considerable study on COVID-19 and the available literature, it appears difficult
to address every element. The article offers a comprehensive examination of
diverse facets concerning the COVID-19 outbreak. It covers a wide range of
topics, such as preventative measures against the virus, clinical
characteristics of symptomatic and asymptomatic individuals, estimations of the
infection and incubation periods, the immune responses that the virus elicits
in humans and the relationship between pre-existing comorbidities and COVID-19
mortality. Furthermore, the article provides a historical framework for understanding
pandemics, tracing their evolution from confined outbreaks to global epidemics,
starting in the 16th and 19th centuries. It delves into zoonotic origins,
elucidating the transmission of zoonoses from animals to humans, with
illustrative examples like HIV/AIDS, Ebola and historical influenza strains. The
SARS-CoV-2 virus exhibits significant genetic similarities to pangolin
coronaviruses and bat betacoronaviruses, indicating that the ongoing COVID-19
pandemic has its origins in an animal reservoir5. As the battle against COVID-19 continues, the
acquisition of knowledge and understanding remains indispensable in formulating
efficacious strategies to safeguard global public health.
2. Emergence and spread of COVID-19 (coronavirus
disease 2019)
Several
pneumonia cases with an unclear cause emerged in late December 2019, in Wuhan,
Hubei Province, China6. The afflicted individuals exhibited
clinical signs of fever, cough, dyspnea, chest pain and bilateral lung
infiltration, symptoms of viral pneumonia, which were comparable to those in
SARS and MERS7. The Huanan Seafood Wholesale Market, a wet
market in Wuhan's downtown known for selling seafood and live animals,
including poultry and wildlife (Figure 1), was linked to the majority of
the initial cases8. On December 8, 2019, the earliest case was recorded9.
Figure 1: The
figure represents the sequence of COVID-19 events recorded. The severe acute
respiratory syndrome coronavirus 2 or SARS-CoV-2, has been identified as the
causative agent of COVID-19 by the International Committee on Taxonomy of
Viruses (ICTV). The World Health Organization (WHO) declared it a public health
emergency of international concern (PHEIC).
World
Health Organization (WHO) was formally notified that the Wuhan Municipal Health
Commission reported an unknown pneumonia outbreak on December 31. Independent
Chinese scientific teams revealed a novel beta coronavirus as the cause of this
newly discovered disease10. The first genome sequence of the novel
coronavirus was made available on January 10. The outbreak coincided with the
Lunar New Year celebrations, which led to more people traveling and spreading
the virus to additional Hubei province cities and ultimately to other regions
of China11. The escalation in severity led the World
Health Organization (WHO) to declare the COVID-19 outbreak as a public health
emergency of international concern on January 3012. WHO officially designated the illness as COVID-19 on
February 1113. (Figure 1) shows the timelines of these
events. China imposed strict public health measures, such as a city-wide
lockdown of Wuhan on January 23 with travel and transportation restrictions, to
contain the outbreak14. The virus's high transmissibility and
global travel contributed to large clusters of infections being reported in
numerous countries. Consequently, on March 11, 2020, the World Health
Organization (WHO) formally declared the COVID-19 outbreak to be a pandemic15,16. China was able to contain the virus quite well, but
cases in the USA and Europe increased drastically17.
3. SARS and COVID-19: Similarities and differences
There are
notable similarities between the clinical manifestations and modes of
transmission of the 2019 COVID-19 and the SARS (severe acute respiratory
syndrome) virus. Both infections have the potential to manifest as rapidly
progressing pneumonia. It seems that the primary mode of transmission for both
is infectious respiratory droplets that are released from mucosal membranes (Table
1). The viruses show comparable stability and degradation in aerosols and
on various surfaces18. According to researchers, both viruses can
live for up to two days on stainless steel and three days on plastic and their
viral titers on both surfaces show comparable decay patterns19-21. Both SARS and COVID-19 seem to have a median incubation
period of 4 to 7 days from first exposure to the start of symptoms.
Furthermore, according to research, the maximum incubation time for both might
be up to 14 days22-24. This longer incubation time adds to the
difficulty of preventing the spread of these illnesses. Despite these
similarities, it is important to emphasize that SARS and COVID-19 are caused by
different viruses and are members of separate coronavirus subfamilies. In
summary, whereas SARS and COVID-19 share clinical signs and transmission
characteristics, they are caused by separate viruses and have distinct
characteristics that distinguish them as distinct causative agents.
Understanding these similarities and differences appears critical for
successful epidemic management and prevention measures. The incubation time and
length of viral shedding are critical for determining the risk of transmission,
adopting isolation and quarantine measures and developing effective antiviral
therapies for patients. According to recent epidemiological studies, the
typical period of COVID-19 virus shedding is around 20 days, with some
survivors shedding for as long as 37 days25. In contrast, viral RNA remained detectable in non-survivors
until death. Severe COVID-19 patients may suffer viral shedding for a median of
31 days after the disease starts26.
Table 1: SARS and COVID-19 comparisons: green highlights
similarities, yellow highlights differences from COVID-19 and orange highlights
feature specific to COVID-19.
|
SARS
COVID-19 | ||
|
Pre-transmissibility |
NO |
YES |
|
Mild
case transmissibility |
NO |
YES |
|
Reproduction
Number |
1.7-1.9
(WHO) |
2.0-2.5
(WHO) |
|
Number
of reported cases |
More
than 8000 |
692.52
million (July 31, 2023) |
|
Number
of reported deaths |
774 |
6,903,467
(July 31, 2023) |
|
Mortality
rate |
9% |
3.1% |
|
The
primary mode of transmission |
Infectious
respiratory droplets dispersed from mucous | |
|
Ability
to survive on surfaces |
YES | |
|
Median
incubation period |
4-7
days | |
|
Maximum
incubation period |
14 days | |
|
Potential
to cause severe respiratory infection |
YES | |
|
Potential
to infect CNS and brain |
YES | |
Betacoronaviruses and alphacoronaviruses have
important natural hosts in bats. RaTG13, a bat coronavirus isolated from Rhinolophus affinis in Yunnan
province, China, is the closest known match to SARS-CoV-2 to date27. RaTG13
and SARS-CoV-2 share 96.2% of the full-length genome sequence, demonstrating a
strong genetic similarity28,29. The fact
that SARS-CoV-2 and RaTG13 share over 90% of their genome's sequence, including
the variable S and ORF8 regions, is especially remarkable28. Their
close relationship is highlighted by phylogenetic analysis, which lends
credence to the theory that bats are the original host of SARS-CoV-2. SARS-CoV-2
and “RmYN02,” a recently discovered coronavirus found in a Yunnan Rhinolophus malayanus bat, share
93.3% of their genome29.
Interestingly, it shares a longer 1ab gene with SARS-CoV-2 with 97.2% identity,
higher than RaTG1330. Furthermore,
ZC45 and ZXC21, two additional bat coronaviruses that were previously
discovered in eastern Chinese Rhinolophus
pusillus bats, are members of the SARS-CoV-2 lineage within the
Sarbecovirus subgenus31. These
findings highlight the wide range of bat coronaviruses that are strongly
associated with SARS-CoV-2, indicating that bats may be the virus's possible
hosts. Recent investigations reveal that the genetic diversity observed in
SARS-CoV-2 and its related bat coronaviruses stems from over 20 years of
sequence evolution. Consequently, it is incorrect to categorize these bat
coronaviruses as the immediate progenitors of SARS-CoV-2, despite being likely
evolutionary ancestors.
Pangolins are another possible animal host
connected to SARS-CoV-2. Between 2017 and 2019, several viruses related to
SARS-CoV-2 were discovered in the tissues of pangolins32. These
pangolin viruses are from two distinct sub-lineages and were independently traced
in the provinces, of Guangxi and Guangdong33-37. Pangolins
linked to various smuggling incidents have been found to have
SARS-CoV-2-related coronavirus infections, suggesting that these animals may
serve as hosts for the viruses38. Pangolins
infected with coronaviruses displayed clinical symptoms and histological
changes such as multiple organ infiltration of inflammatory cells and
interstitial pneumonia, in contrast to bats, which typically carry the virus
without obvious damage39.
Emerging coronaviruses that are derived from bats
require an intermediate host to proliferate. For example, dromedary camels and
palm civets served as intermediary hosts for SARS-CoV and MERS-CoV, respectively40. The
viruses harboured by these hosts share a genome sequence identity of over 99%
with the corresponding viruses in humans41. The role
of an intermediary host in the transmission of the SARS-CoV-2 virus, which is
accountable for the COVID-19 pandemic, is under scrutiny and remains unclear.
Pangolin coronaviruses exhibit only a 92% genomic identity with SARS-CoV-2,
despite displaying a remarkably similar receptor-binding domain (RBD)42.
Consequently, it is challenging to definitively ascertain whether pangolins
acted as the intermediate host for SARS-CoV-2 or if they were directly
implicated in the virus's emergence. The animal source of SARS-CoV-2 is
presently poorly understood, with limited knowledge available on this aspect.
The virus's reservoir hosts have not been identified, nor it has been
determined if an intermediate host was involved in the virus's transmission to
humans. Significantly, the discovery of pangolin coronaviruses, RaTG13 and
RmYN02 implies that SARS-CoV-2-like coronaviruses are prevalent in animals43-44.
In addition to wildlife, research has explored the
susceptibility of domesticated and laboratory animals to SARS-CoV-2 infection.
Experimental findings have demonstrated that SARS-CoV-2 can effectively
replicate in cats and ferrets, particularly in the upper respiratory tract.
Conversely, dogs, pigs, chickens and ducks have exhibited immunity to the virus45. Notably,
minks have been observed to contract SARS-CoV-2, as evidenced by an outbreak on
mink farms in the Netherlands, leading to severe cases of respiratory distress
and interstitial pneumonia46. Although
devoid of symptoms, two dogs in Hong Kong tested positive for spontaneous
SARS-CoV-2 infection through serological and virological tests47. Similarly,
blood samples from cats in Wuhan showed the presence of neutralizing antibodies
against SARS-CoV-2, confirming the infection in cat populations. However, the
possibility of transmission from cats to humans is still uncertain48. Ongoing
comprehensive research and surveillance on animal susceptibility aim to provide
a deeper understanding of potential hosts and transmission dynamics of the
virus.
4. Comparative insights into SARS-CoV-2:
Infectiousness, transmission and evolution
The virus
accountable for acute respiratory illness, SARS-CoV-2, belongs to the
coronavirus family and carries a non-segmented genome composed of
positive-sense, single-stranded RNA enveloped by the viral capsid49. Coronaviruses (CoVs) are categorized into four genera:
α, β, γ and δ-CoV50. While α- and β-CoV predominantly infect
mammals, they can also affect birds. Human-infecting coronaviruses include
HCoV-229E, SARS-CoV, HCoV-OC43, HCoV-NL63, MERS-CoV and HCoV-HKU151. Infections caused by HCoV-229E, HCoV-NL63, HCoV-HKU1
and HCoV-OC43 typically result in mild respiratory symptoms, whereas SARS-CoV
and MERS-CoV can lead to severe respiratory disease, occasionally resulting in
death due to multiple organ failure52. SARS-CoV-2 shares notable similarities (over 85%) with
bat-derived SARS-like coronaviruses identified as bat-SL-CoVZC45 and
bat-SL-CoVZXC2153. In comparison to SARS-CoV and MERS-CoV, it demonstrates
approximately 79% and 50% homology, respectively54. This evidence, combined with phylogenetic research,
strongly indicates that SARS-CoV-2 originated in bats and potentially
transmitted to humans through an unidentified intermediate host species. (Figure
2) illustrates the genomic structure, encoded structural and non-structural
proteins and the primary host of SARS-CoV-2.
The
pathogenesis of SARS-CoV-2 involves a complex interplay of viral and host
factors. As an enveloped positive-sense single-stranded RNA virus, the genomic
structure of SARS-CoV-2 comprises a significant portion (two-thirds) dedicated
to an open reading frame (ORF 1a/b), encoding 16 non-structural proteins (NSPs)
crucial for replication. The remaining section of the genome encodes essential
structural proteins (Spike glycoprotein, Small Envelope protein, Matrix protein
and Nucleocapsid protein) and accessory proteins with functions still under
investigation. The S glycoprotein, essential for host cell entry, binds to the
angiotensin-converting enzyme 2 (ACE2) receptor. However, the precise mechanism
of membrane invagination for SARS-CoV-2 endocytosis remains unclear. Host
factors, particularly ACE2 expression, influence viral tropism. The elderly and
individuals with underlying health conditions are more susceptible to severe
infections, partly due to age-related immune system changes and comorbidities.
Host immune responses, both innate and adaptive, play a crucial role and
dysregulated responses can contribute to disease severity. Additionally,
genetic factors contribute to interindividual variability in susceptibility and
disease outcomes. A comprehensive understanding of these viral and host
elements is crucial for developing effective therapeutic interventions and
vaccines against SARS-CoV-2. Ongoing research continues to unveil additional
details about the intricate virus-host interactions shaping the pathogenesis of
COVID-19. Without a doubt, expressive experimentation has shown that the virus
infects people by attaching itself to respiratory system-expressed ACE2
receptors55,56. Overall, findings from several
investigations show that SARS-CoV-2 is extremely infectious, with viral
shedding commencing before symptoms develop and the virus spreading through
many channels. Controlling the disease's spread is a key problem for public
health initiatives.
SARS-CoV-2
is less severe in terms of morbidity and mortality than MERS and SARS, but it
is more contagious. COVID-19 has a much lower mortality rate of 3.4% compared
to 9.6% and 35% for SARS and MERS, respectively. COVID-19 primarily spreads
through person-to-person contact, particularly between close friends and family
members57. Numerous studies demonstrate the critical
role symptomatic individuals play in COVID-19, mainly through respiratory
droplet expulsion from actions like coughing or sneezing. On the other hand,
nosocomial transmission was primarily responsible for the spread of MERS-CoV
and SARS-CoV among healthcare personnel58. In MERS-CoV outbreaks, medical staff was responsible
for 62%-79% of cases, whereas in the SARS case, they accounted for 33%-42% of
cases. The most likely ways for a virus to spread are through direct contact
with the host or interactions with an unidentified intermediate carrier59.
Figure 2: The
figure represents the structural and genetic characteristics of SARS-CoV-2.
Numerous structural proteins, including the spike glycoprotein (S), envelope
(E), matrix (M) and nucleocapsid protein (N). The genetic segment ORF1ab
encodes several non-structural proteins (nsp 1–16) concurrently. The
host-related variables that can affect an individual's susceptibility to and
the severity of a SARS-CoV-2 infection are listed in the lower section.
The
SARS-CoV-2 virus changes in a variety of ways as it grows and spreads among people.
In December 2020, a noteworthy variant, VUI-202012/01, was examined due to 17
distinct alterations or mutations in its DNA. Since the discovery of the
SARS-CoV-2 virus in 2019, thousands of mutations have already manifested in its
genome60. As the pandemic continues, the continual mutation process
in the population may result in the production of immunologically relevant
mutations, thereby affecting vaccination effectiveness. These mutations are
resulting in novel viral combinations. The COVID-19
genomics UK consortium (COG-UK)
has conducted extensive epidemiological and virological investigations in
response to the significant surge in COVID-19 cases recently observed in the
United Kingdom (UK), particularly in South East England61. A novel variant was identified in viral genome sequences, forming a
distinct phylogenetic grouping. This variant is distinguished by multiple spike
protein mutations (deletion 69-70, deletion 144, N501Y, A570D, D614G, P681H,
T716I, S982A and D1118H), accompanied by alterations in other genomic regions62. Although viral mutations are normal, preliminary studies show that
this variant in the UK may be critical for increased transmissibility and is
projected to possibly raise the reproductive number by 0.4 or more63. Notably, this new variety evolved during a period of increased family
and social gatherings. However, there is no indication that it causes more
severe infections than other variations.
5. Role of ACE-2 Receptor, an Angiotensin-Converting
Enzyme
SARS-CoV-2
gains entry into the human host through receptor-mediated endocytosis, a
mechanism where viruses bind to specific receptors on the host cell's surface,
facilitating entry. The virus's receptor-binding domain establishes a
connection with the appropriate receptor on the host cell, enabling entry. Both
SARS-CoV and SARS-CoV-2 utilize the angiotensin-converting enzyme-2 (ACE-2)
receptor to infect cells. Earlier studies have shown that the S-protein of
SARS-CoV exhibits a strong affinity for the ACE-2 receptor, serving as the
entry point for the virus into host cells64. Figure 3 depicts the fusion of the virus with the host
receptor.
The entry
of SARS-CoV-2 into host cells is also mediated by S-protein priming by transmembrane
protease serine-2 (TMPRSS2). This priming event is crucial for the fusion of
the viral envelope with the host cell membrane, enabling subsequent viral
entry. Therefore, the coordinated interplay between the ACE-2 receptor and
TMPRSS2 is essential for the efficient entry of SARS-CoV-2 into the host
environment. It is noteworthy that TMPRSS2 exhibits higher expression and
broader distribution compared to ACE-2 receptors, suggesting that ACE-2 may act
as a limiting factor during the initial infection phase. While TMPRSS2 is a key
component for viral entry, alternate proteases, such as cathepsin B/L, may act
as substitutes for TMPRSS2. Hence, the simultaneous inhibition of these
proteases becomes crucial in preventing cellular entry.
The
structural characteristics of the S-proteins of SARS-CoV and SARS-CoV-2
facilitate the entry of the latter into cells65. Studies involving human HeLa cells and animals with and
without ACE-2 expression support the involvement of ACE-2 receptors in the
cellular entry of the SARS-CoV-2 virus, particularly the Wuhan strain66. Research on SARS-CoV-2 infection of BHK21 cells
indicated higher infection rates when transfected with human and bat ACE-2
receptors compared to BHK21 cells lacking ACE-2 expression67-69. Biophysical and structural data suggest that the ACE-2
binding affinity of the SARS-CoV-2 S-protein ectodomain is significantly
greater than that of the SARS-CoV S-protein by a ratio of 10:2070. This difference is believed to contribute to the
variance in contagiousness between SARS-CoV-2 and SARS-CoV. Although the ACE-2
and ACE-1 receptors share similarities, the ACE-2 receptor has a smaller active
site and a smaller binding pocket with different amino acids, making it
resistant to typical ACE inhibitors such as lisinopril, enalapril and ramipril71.
Furthermore,
there is no evidence suggesting that angiotensin receptor blockers (ARBs), like
losartan, disrupt the activity of ACE-2. TMPRSS2, identified as a type II
transmembrane protease, consists of distinct domains, including an
intracellular N-terminal domain, a transmembrane domain, an extracellularly
extending stem region and a C-terminal domain facilitating its serine protease
(SP) function72. The serine protease activity relies on a
catalytic triad, comprised of His296, Asp345 and Ser441, responsible for
cleaving basic amino acid residues, particularly lysine or arginine residues,
aligning with its role in cleaving the S1/S2 site in SARS-CoV-273.
While
TMPRSS2 has been recognized for its involvement in prostate cancer and viral
infections such as influenza, SARS and MERS, it has recently gained attention
from drug developers. Multiple studies are underway to uncover strategies aimed
at reducing TMPRSS2 expression or blocking its activity in host cell membranes,
with the ultimate goal of inhibiting SARS-CoV-2 entry into host cells74,75.
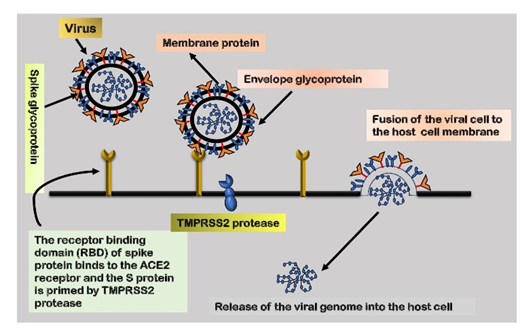
Figure 3: A
representation of the SARS-CoV-2 spike protein with the host receptor and the
subsequent fusion of the viral cell with the host cell membrane.
6. Diagnostic, Therapeutic Approaches and
Strategies to Inhibit Viral Entry
Molecular
detection of SARS-CoV-2 nucleic acid is the most accurate diagnostic approach72. Various commercially available kits for viral nucleic
acid detection target different genes, including ORF1ab (containing RdRp), N, E
or S73. The detection time may vary from a few
minutes to several hours depending on the technology utilized. Although
SARS-CoV-2 has been detected in throat swabs, posterior oropharyngeal saliva,
nasopharyngeal swabs, sputum and bronchial fluid, the virus load is notably
higher in samples from the lower respiratory tract74. Viral nucleic acid has also been detected in intestine and
blood samples, even in cases where respiratory tests yielded negative results.
The viral load may decrease from its peak at the onset of the illness,
potentially leading to false negatives when using oral swabs75. It is advisable to employ multiple detection techniques
to confirm a COVID-19 diagnosis.
To
address the issue of false negatives, alternative detection approaches have
been utilized. Therefore, for individuals with a robust clinical suspicion of
COVID-19 despite an initial negative nucleic acid screening, a combination of
CT scans and repeated swab testing has been recommended. Serological assays
that identify antibodies to the N or S protein of SARS-CoV-2 could complement
molecular diagnosis, particularly in the latter stages of the illness or for
retrospective research76,77. The magnitude and duration of immunological
responses are still unknown and the sensitivity and specificity of existing
serological assays vary. When choosing and interpreting serological testing,
all of these factors should be taken into consideration, possibly even
extending to future assays for T-cell responses.
As of
right now, neither COVID-19 nor specific antivirals that target SARS-CoV-2 have
the potential to combat the disease. However, several treatments have shown
some promise. Manufacturers and researchers are undertaking large clinical
studies to examine new COVID-19 therapy options. As of October 2, 2020, over
405 therapeutic medicines were being developed for COVID-19, with almost 318 of
them undergoing human clinical trials78. Potential antiviral target for the treatment of
COVID-19 is depicted in (Figure 3).
A crucial
strategy in combatting SARS-CoV-2 infection is to hinder viral entry.
Angiotensin-converting enzyme 2 (ACE2) exists in membrane-bound ACE2 (mACE2),
located in the gallbladder, heart, intestines, kidneys and testes79. The virus uses human proteases as entry activators to
break through host cells through membrane fusion and it uses ACE2 as a
receptor. Treatments aimed at this entry mechanism have the potential for
treating COVID-19. Umifenovir, also known as arbidol, is a drug approved for treating
respiratory viral infections and influenza in China and Russia. Its mechanism
of action involves preventing membrane fusion by interfering with the
interaction between the S protein and ACE279. In vitro studies have demonstrated its efficacy against
SARS-CoV-2, Clinical data suggests that it may present a more effective
treatment for COVID-19 when compared to lopinavir and ritonavir80-84.
One
notable drug that shows promise is camostat mesylate, which is licensed in
Japan to treat postoperative reflux esophagitis and pancreatitis85. Previous studies have demonstrated the ability of
camostat mesylate to inhibit TMPRSS2 activity and protect mice from fatal
SARS-CoV infection86,87. Recent studies have further indicated that
camostat mesylate can inhibit the entry of SARS-CoV-2 into human lung cells88. This suggests potential utility as an antiviral drug
against SARS-CoV-2 in the future, although further clinical data is required to
confirm its effectiveness.
Other
drugs used to treat autoimmune diseases and prevent malaria, such as
chloroquine and hydroxychloroquine, may also influence SARS-CoV-2 entry. They
work by preventing membrane fusion by raising endosomal pH, interfering with
the interaction between virus and host receptor and inhibiting the
glycosylation of cellular receptors89-91. Regarding their effectiveness in treating COVID-19, there
remains a lack of scientific consensus. Despite concerns about an increased
risk of cardiac arrest in treated patients, two clinical investigations found
no correlation between these medications and patient mortality rates92,93. On June 15, 2020, due to documented adverse events, the
US Food and Drug Administration (FDA) revoked the emergency use authorization
for chloroquine and hydroxychloroquine in COVID-19 therapy94.
Another
therapeutic approach involves the use of soluble recombinant hACE2, specific
monoclonal antibodies or fusion inhibitors targeting the SARS-CoV-2 S protein
to prevent its binding to the ACE-2 receptor95. Examples of replication inhibitors include remdesivir
(GS-5734), favilavir (T-705), ribavirin, lopinavir and ritonavir. The remaining
three agents act on RdRp, except for lopinavir and ritonavir, which inhibit
3CLpro. (Figure 4) illustrates potential antiviral targets for COVID-19
treatment. However, further clinical research is necessary to evaluate the
effectiveness and safety of these approaches.
Figure 4: Potential
antiviral interventions against the SARS-CoV-2. In addition to antiviral agents
immunomodulatory and immunoglobulin-based medications are potential treatments.
Key molecular targets implicated in the viral replication cycle and potential
treatments include ACE2 (Angiotensin-Converting Enzyme 2), crucial for the
virus's initial interaction during receptor binding; 3CLpro (3C-Like Protease),
a protease inhibited by lopinavir and ritonavir; CR3022, a human monoclonal
antibody targeting the SARS-CoV virus; Envelope Protein (E), a potential target
for disrupting viral replication; Endoplasmic Reticulum (ER), involved in
various stages of viral replication and a potential therapeutic target; gRNA
(Genomic RNA), a critical component of the viral replication process; HR2P
(SARS-CoV-2 Spike Protein Derived Peptides, Hepatod Repeat 2), peptides
considered for their potential in inhibiting viral fusion; ISG
(Interferon-Stimulated Gene), targeted by immunomodulatory agents to modulate
the host immune response; M (Membrane Protein), a potential target for
disrupting viral replication; RNA-Dependent RNA Polymerase or RdRp, the key
enzyme in the viral replication process targeted by antiviral agents such as
remdesivir, favilavir and ribavirin; sgRNA (subgenomic RNA), involved in
various stages of the replication cycle; S (Spike Protein), a major target for
therapeutic intervention considering its role in receptor binding and viral
entry; and TMPRSS2 (Transmembrane Protease Serine Protease 2), facilitating
viral entry into host cells and a potential target for antiviral strategies.
7. Current Management approaches for COVID-19
Avoiding
transmission should be the primary objective of COVID-19 treatment, especially
in those with moderate symptoms, given the uncertainty surrounding the
effectiveness of currently available antiviral medications. Individuals
receiving at-home care must be closely monitored and if their health worsens,
therapy must be escalated right away. Studies on the advantages of
corticosteroids, weighing anti-inflammatory effects with possible hazards of
viral replication, have shown conflicting findings96. Corticosteroids may, however, be taken into
consideration in situations when there are other signs, such as severe COPD.
Inhalers are used over nebulized medicines, which produce aerosols, to reduce
the danger of airborne viral dissemination97. Nonsteroidal anti-inflammatory drugs (NSAIDs) have
generated controversy because of their ability to affect epithelial cell ACE2
receptor levels and perhaps worsen viral infection98. The specific effects of NSAID usage in COVID-19 remain
uncertain. Some suggest that NSAIDs might elevate the risk of acute respiratory
distress syndrome (ARDS) by triggering leukotriene release and
bronchoconstriction99. However, the application of NSAIDs for
symptom management should be tailored to each individual. Presently, the European
Medicines Agency (EMA) and the World Health Organization (WHO) do not advise
against the use of NSAIDs100. In hospital settings, acetaminophen is
often preferred over NSAIDs to minimize the risk of bleeding and kidney damage101.
Controversy
has arisen regarding the use of angiotensin receptor blockers and ACE
inhibitors in COVID-19. Nonetheless, the American Society of Cardiology and the
European Society of Cardiology presently do not recommend initiating or
discontinuing these drugs102. The choice of antiviral and
anti-inflammatory therapies should be personalized according to each patient's
situation, guided by infectious disease experts and conducted within the
context of a clinical trial or registry. Oxygen therapy, encompassing methods
such as nasal cannula and high-flow oxygen, is often beneficial for individuals
with mild to severe COVID-19103. Non-invasive and invasive mechanical
ventilation are commonly needed in situations of acute respiratory failure.
Positive airway pressure (PAP) is an aerosol-generating treatment; hence
healthcare professionals had and must use a greater degree of personal
protective equipment (PPE)104. Unless there are particular
contraindications, pharmaceutical prophylaxis was used for these events and should
be made available to hospitalized COVID-19 patients due to the elevated risk of
venous thromboembolism.
8. Preventing COVID-19: Progress in vaccine advancements
Nearly
200 clinical studies were conducted to evaluate a range of innovative and
repurposed medicines in the battle against COVID-19105. Among these programs, vaccinations hold promise since they
could stop the spread of illness to a larger population. Before they may be
used widely, the safety and efficacy of these immunizations must first be
properly confirmed. It is impossible to overstate the importance of this stage
since subpar immunizations run the danger of doing more damage than good via
mechanisms including antibody-dependent augmentation. Therefore, meticulous
testing and verification are crucial before the widespread adoption of any
COVID-19 immunizations.
8.1. Technological approaches employed in the
development of COVID-19 vaccines
Numerous
technologies, were employed by scientists and researchers globally in their endeavours
to create a secure and efficient vaccine for SARS-CoV-2. Among these
technologies, gene vaccines, inactivated vaccines, viral vector vaccines and
protein subunit vaccines stand out as the most promising candidates.
8.2. Vaccines based on protein subunits
Protein
subunit vaccines, frequently administered through sophisticated systems like
liposomes, virosomes or polymeric nanoparticles, harness components of the
pathogen to stimulate the host's immune system105. Liposomes and virosomes, serve as effective adjuvants
and carriers for antigens and are commonly employed in the development of
vaccines against SARS-CoV-2106. For instance, a study reflects upon a cationic
liposome protein subunit vaccine that incorporates the S1 component of the
SARS-CoV-2 virus. This vaccine also includes two adjuvants: monophosphoryl
lipid A (MPLA), acting as a TLR4 and TLR9 agonist and CpG ODN107. The inclusion of cationic elements such as
1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) enhances the interaction of
the liposome with antigen-presenting cells108. This liposome vaccine demonstrated improved T cell
immunity, activating CD4+ and CD8+ cells and promoting
IgA synthesis for potential mucosal defense109.
Virosomes,
lipid vesicles containing viral proteins, are preferred over liposomes as
adjuvants due to their ability to shield pharmaceutically active compounds from
degradation in endosomes until they reach the cytoplasm110. Virosomes have previously been utilized in the delivery
of vaccines for SARS-CoV and MERS-CoV. The Centre for Vaccine Development at
Texas Children's Hospital, Baylor College of Medicine, is working on a subunit
vaccine against SARS-CoV-2. This vaccine employs a recombinant S-protein
receptor-binding domain (RBD), likely combined with alum or glucopyranosyl
lipid A (GLA), a synthetic TLR4 agonist111.
The Australia
University of Queensland and Novavax collaborated on the development of an
immunogenic virus-like nanoparticle vaccine, NVX-CoV2373, currently in phase 3
trials. This vaccine incorporates a recombinant S-protein, demonstrating
minimal reactogenicity and eliciting a T helper 1 response without significant
side effects in most individuals112. Clover Biopharmaceuticals is also working on a highly
pure S-trimer vaccine using their Trimer-Tag technology, previously employed in
subunit vaccines for HIV, RSV and Influenza. In collaboration with
GlaxoSmithKline (GSK) and Dynavax Technologies, Clover Biopharmaceuticals has
completed enrolment in a phase 1 study, employing the CpG 1018 adjuvant, a TLR9
agonist known to activate CD4+ and CD8+ T cells with a
favorable safety profile113,114.
8.3. Vaccines with inactivated viruses
Weakened
bacterial or viral pathogens used in inactivated vaccinations stimulate the
immune system without actually infecting the recipient. Although these
vaccinations don't provide lifelong protection, booster injections are often
required to provide a long-term shielding effect. Large numbers of viral
particles are propagated, condensed and then made inactive using chemical
and/or physical techniques to make inactivated viral vaccines. Various
techniques, such as the application of ascorbic acid, binary ethylenimine,
gamma irradiation and high-temperature treatment, are commonly employed to
render viral particles inactive115. The efficacy of these approaches relies on ensuring
complete deactivation of the specific virus. The Wuhan Institute of Biological
Products, affiliated with the China National Pharmaceutical Group (Sinopharm),
actively worked on one of the initial inactivated COVID-19 vaccines116,117. In the development of this vaccine, the virus undergoes
growth in the Vero cell line, followed by inactivation using formalin or
β-propiolactone, with alum incorporated as an adjuvant118. All participants in the phase 1/2 clinical trials
developed antibodies in response to the vaccination, with few negative side
effects119.
The most
typical adverse effects, such as discomfort at the injection site and fever,
were modest and self-limiting. Phase 3 studies are now being conducted to
assess the vaccine's effectiveness and long-term safety. Sinovac Biotech Ltd.
in China is involved in developing CoronaVac (formerly PiCoVacc), another
inactivated vaccine. This vaccination puts genetic stability first, using the
SARS-CoV-2 CN2 strain that was isolated from bronchoalveolar lavage fluid samples
of hospitalized patients. The vaccine is presently in the midst of phase 3
clinical trials, involving a participant pool of 8,870 individuals120,121. In a distinct development, the University of Wisconsin,
Madison, has collaborated with vaccine companies FluGen and Bharat Biotech to
create an inactivated vaccine named CoroFlu, designed for intranasal delivery.
Derived from FluGen's M2SR influenza vaccine, CoroFlu leverages the immune
response targeting influenza. The M2SR vaccine has been adapted to incorporate
the S-protein gene sequences of SARS-CoV-2, to elicit an immune response
against the virus122. This non-invasive nasal immunization
approach shows potential in eliciting robust mucosal and systemic immune
responses to combat respiratory virus infections, providing an alternative to
traditional invasive parenteral vaccination methods.
8.4. Adenovirus-based COVID-19 vaccines
Adenoviruses,
with their icosahedral capsid and double-stranded linear DNA, are essential for
initiating both innate and adaptive immunity in mammals. By increasing
cytotoxic T lymphocytes and releasing pro-inflammatory cytokines, they aid in
the immune response. These lymphocytes are in charge of identifying and getting
rid of virus-infected cells123. Building on this method, adenoviral vectors
have been extensively employed to combat a variety of illnesses, including
influenza, Ebola, SARS, HIV and recently COVID-19124. Renowned academic institutions and pharmaceutical
companies including the Jenner Institute at Oxford University, CanSino
Biologics and Johnson & Johnson have led the development of COVID-19
vaccines utilizing adenoviral vectors125. Phase 2 clinical trials for CanSino Biologics' Ad5-nCoV
vaccine are presently underway and the results are promising126. This vaccine carries the genetic code for the S-protein
of the SARS-CoV-2 virus and employs the non-replicating chimpanzee adenoviral
vaccine vector, AZD1222. Noteworthy is its suitability for vulnerable
populations such as children, the elderly and individuals with pre-existing
medical conditions, as it necessitates only a single dose and triggers a
substantial immune response without causing illness127. AstraZeneca and the University of Oxford have conducted
phase 1 and phase 2 studies on AZD1222, demonstrating a promising safety
profile and the successful generation of neutralizing antibodies against
SARS-CoV-2128,129.
Adenoviral
vectors are still in the early stages of development and have not yet been
approved for use in the treatment of infectious diseases in humans, even though
they exhibit great promise for COVID-19 vaccines. Concerns have been raised
about possible inflammatory responses as reported in AstraZeneca studies. Additionally,
it's probable that people already have some amount of resistance to adenoviral
vectors owing to their frequent exposure to them. While research and clinical
trials continue, the scientific community is dedicated to developing safe and
effective medicines to combat the COVID-19 pandemic and the long COVID.
8.5. Nucleic acid-based vaccines
DNA
vaccines or mRNA vaccines, promise to be more effective than conventional immunizations.
Direct administration of DNA plasmids that encode particular target antigens
results in potent B and T cell responses with increased safety130. These vaccinations are safe for those with impaired
immune systems since they don't include any infectious organisms. Synthetic DNA
vaccines quicken the development process by enabling scalable manufacture, fast
design and preclinical testing of several candidates and simpler regulatory
approval for clinical use. Their stability at different temperatures also
guarantees a longer shelf life. Currently being developed is a gene-based
vaccine that specifically targets the S-protein of SARS-CoV-2. The vaccine
candidate from Inovio Pharmaceuticals uses DNA-plasmid pGX9501, which was
developed using MERS-CoV vaccine constructions from the past. The vaccine is
now in phase 2 clinical trials. It is given intradermally and then
electroporated131. Gene vaccines also use mRNA, which operates
in the cytoplasm without having to cross the nuclear membrane, in addition to
DNA. mRNA vaccines are less dose-intensive than DNA vaccinations and produce
strong immune system memory. However, because of their heat lability and
susceptibility to hydrolysis by circulating ribonucleases, they are less stable132. This is addressed by the formulation of mRNA vaccines
as lipid nanoparticles, which improve stability and host distribution. Examples
include the SARS-CoV-2-targeting drugs mRNA-1273 from Moderna and BNT162b1 from
Pfizer, both of which are in advanced clinical moderation133. Despite the advancements, there are still difficulties
in the global production, distribution and administration of COVID-19 vaccines.
8.6. Drugs approved for the COVID-19 treatment
Ongoing
extensive clinical trials are underway to evaluate the potential effectiveness
of several medications in treating COVID-19 patients. The selection of these
medications is based on the hypothesis that they may hinder the virus from
entering the host and replicating. Various compounds, including some that have
undergone human clinical trials, are currently under assessment in clinical
trials as potential COVID-19 treatments. Researchers are investigating the
ability of experimental drugs to impede the virus's entry into the host and
subsequent replication. While certain medications have been previously employed
in treating SAR-CoV infections, others are being utilized for the first time in
the context of SARS-CoV-2 infections.
Remdesivir,
developed by Gilead Sciences, has received FDA approval for treating COVID-19
in patients aged 12 and above requiring hospitalization. Remdesivir works by
inhibiting the RNA-dependent RNA polymerase, disrupting its interaction with
the RNA of SARS-CoV-2 and thereby halting further replication134. After receiving remdesivir intravenously, 36 out of 53
COVID-19 patients showed improvement, indicating positive clinical outcomes135. Although lopinavir and ritonavir-based antiretroviral
therapy have been investigated, it has not proven to be any more effective than
standard care. Umifenovir, which has been licensed for influenza prevention in
China and Russia, is used for the treatment of COVID-19 because of its
potential to inhibit the S-protein/ACE2 interaction136.
Research
indicates that favipiravir, which inhibits RNA polymerase and is approved for
use against influenza in Japan, has a better clinical outcome in mild cases of
COVID-19 than umifenovir137. In small-scale clinical trials conducted in
China, chloroquine has demonstrated potential in slowing pneumonia progression
and viral replication in COVID-19 patients138-139. For COVID-19 patients, there is promise in the combination
of statins and angiotensin receptor blockers (ARBs) to prevent acute
respiratory distress syndrome (ARDS)140. Ongoing studies are exploring the potential benefits of
these combined treatments in managing the severe consequences of the illness (Figure
5). The strategy of employing existing, approved drugs for COVID-19
treatment capitalizes on the current pharmacopeia to swiftly address the urgent
global health crisis. This tactic comprises repurposing well-known
pharmaceuticals that were first authorized for a range of medical conditions to
target particular aspects of the SARS-CoV-2 virus or the host immune system.
Making the most of these medications' well-established pharmacokinetics,
mechanisms of action and safety profiles is the main goal.
Remdesivir,
initially developed for Ebola, is repurposed as an antiviral for COVID-19. Its
mechanism of action involves inhibiting the viral RNA polymerase, thereby
disrupting viral replication141. Lopinavir/Ritonavir, FDA-approved for
treating HIV, is being explored for its ability to inhibit the 3CLpro enzyme in
SARS-CoV-2, disrupting viral replication142. Agents with anti-inflammatory and immunomodulatory
properties, such as dexamethasone-a potent corticosteroid with strong
anti-inflammatory effects-are being repurposed to alleviate the severe
inflammatory responses observed in critically ill COVID-19 patients,
potentially reducing mortality143. The anti-inflammatory characteristics of azithromycin,
an antibiotic, are currently under investigation for their ability to regulate
the immune system and mitigate inflammation in individuals with COVID-19144. Monoclonal antibodies, designed to specifically target
SARS-CoV-2, are hypothesized to neutralize the virus, offering targeted
therapeutic intervention.
Convalescent
plasma, derived from individuals who have successfully recovered from COVID-19,
contains antibodies that may neutralize the virus in infected patients, thereby
enhancing the host's immune response145. Antibiotics and antiparasitic agents, including
ivermectin, known for their well-established safety profile, are undergoing
examination for potential antiviral effects against SARS-CoV-2. Additionally,
azithromycin, an antibiotic, is explored for its potential synergy with other
treatments in COVID-19 cases146.
Figure 5: The
illustration outlines the potential steps involved in the entry and replication
of SARS-CoV-2. The sequence initiates with the conformational change of the
viral S protein, triggered by its binding to the cellular ACE2 receptor. This
interaction facilitates the fusion of the viral envelope with the cell membrane
through the endosome pathway. The genomic RNA undergoes translation, leading to
the synthesis of the viral replicase polyproteins pp1a and 1ab. Viral proteases
then cleave these polyproteins, generating smaller functional products.
Following this, the viral polymerase transcribes irregularly, resulting in the
production of subgenomic mRNAs. These subgenomic mRNAs, in turn, contribute to
the translation of different viral proteins. During the assembly phase, viral
proteins and genomic RNA combine to form virions within the endoplasmic
reticulum (ER) and Golgi apparatus. The ER-Golgi intermediate compartment
(ERGIC) plays a pivotal role in the maturation and transportation of virions.
Ultimately, assembled virions are encapsulated into vesicles and released from
the host cells.
9. Challenges and prospects
One major
challenge is the ongoing emergence of new SARS-CoV-2 virus variants. These
variants may acquire increased transmissibility, be resistant to immunity from
previous infections or vaccines and may lead to more severe disease. Monitoring
and adapting to these variants will be an ongoing challenge. Ensuring equitable
and efficient distribution of COVID-19 vaccines in itself remains one of the
major challenges. Disparities in access to vaccines can exacerbate the global
health crisis and hinder efforts to achieve herd immunity. Vaccine hesitancy
and misinformation continue to impede vaccination efforts. Promoting vaccine
education and addressing concerns is crucial to achieving widespread
vaccination and ending the pandemic. The long-term health effects of COVID-19,
also referred to as “long COVID,” are still difficult to understand. Some
individuals experience persistent symptoms and complications long after
recovering from the acute phase of the disease147,148. Healthcare systems in many regions around the globe are
still grappling with the strain of the pandemic. Treating severe cases of
COVID-19 can overwhelm hospitals and lead to delays in providing care for other
serious medical conditions. The pandemic has caused severe economic and social unrest,
Global cooperation and coordination are necessary to combat the pandemic
effectively.
Research
and development of booster shots and updated vaccines will likely continue to
address emerging variants and provide longer-lasting immunity. The development
of effective antiviral drugs to treat COVID-19 may improve outcomes for those
infected and reduce the severity of the disease. Achieving herd immunity
through vaccination remains a key goal for ending the pandemic. Encouraging
vaccination in underserved communities and improving vaccine access are
essential components of this effort149. The experience with COVID-19 underscores the need for
improved pandemic preparedness, early warning systems and global response
mechanisms to mitigate the impact of future infectious disease outbreaks. The
pandemic has accelerated the adoption of telemedicine and digital healthcare
solutions150. These innovations may continue to transform
healthcare delivery and improve access to medical care. Addressing the mental
health challenges arising from the pandemic will be a long-term prospect.
Investing in mental health services and support systems is crucial for
recovery. Promoting good hygiene habits and raising public health awareness can
be very effective in stopping the transmission of contagious illnesses like
COVID-19.
10. Conclusion
In
conclusion, COVID-19 presents a range of challenges, but there is also hope for
the future. Effective vaccination, treatments, global cooperation and
preparedness efforts can contribute to bringing the pandemic under control and
better preparing the world to respond to future health crises. The
comprehensive review provides an in-depth and enlightening examination of the
COVID-19 pandemic through September 2021. Millions of people have been affected
by the SARS-CoV-2 pandemic, which has created previously unheard-of challenges
for global health. The review article elaborates on
several aspects of the illness, starting with its zoonotic origin and moving on
to person-to-person transmission, as well as a map of its geographic
distribution across continents. An important subject included in this research
is COVID-19's clinical manifestations, which may vary from modest respiratory
symptoms to severe instances leading to pneumonia, ARDS and multi-organ
failure. Examining the disease's impact on different age groups and vulnerable
populations, the article highlights the need for specialist healthcare
strategies to protect those who are most susceptible. The report also
discusses several COVID-19 diagnostic methods, including molecular tests like
PCR and antigen assays and serological testing for detecting antibodies. These
tests are necessary for controlling illnesses, tracking contacts and
establishing containment procedures. Investigated in the search for effective
therapies include repurposed drugs, immunomodulatory therapy and antiviral
drugs. The development and administration of vaccines are seen as crucial
strategies for halting the pandemic and promoting herd immunity. The article
acknowledges that COVID-19 comprehension and management have advanced
significantly, but challenges and hurdles still wait for the global healthcare
systems. The challenges include handling viral alterations understanding novel
varieties, combating false information and vaccination resistance and becoming
ready for impending outbreaks. The review provides a basis for further research
and information for public health activities and scientists working at the
molecular level, helping to decrease the consequences of the epidemic and
prepare for any future health crises.
11. Abbreviations
COVID-19: Coronavirus disease 2019; SARS-CoV-2: Severe Acute
Respiratory Syndrome Coronavirus 2; ARDS: Acute Respiratory Distress Syndrome;
PCR: Polymerase Chain Reaction; HIV/AIDS: Human Immunodeficiency Virus/Acquired
Immunodeficiency Syndrome; MERS-CoV: Middle East Respiratory Syndrome Coronavirus;
ICTV: International Committee on Taxonomy of Viruses;
WHO: World Health Organization; PHEIC: Public Health Emergency of International
Concern; RNA: Ribonucleic Acid; CNS: Central Nervous System; RBD: Receptor-binding
Domain; RBM: Receptor-binding Motif; S: Spike Glycoprotein; E: Envelope; M: Matrix;
N: Nucleocapsid Protein; ORF1ab: Open Reading Frame 1ab; nsp: Non-structural Protein;
DNA: Deoxyribonucleic Acid; COG-UK: COVID-19 Genomics UK Consortium; TMPRSS2: Transmembrane
Protease Serine 2; ACE-2: Angiotensin-converting Enzyme 2; SP: Serine Protease;
S1/S2 site: Spike Protein Cleavage site; RdRp: RNA-dependent RNA polymerase;
CT: Computed Tomography; ACE2: Angiotensin-converting Enzyme 2; T-cell:
T-lymphocyte; SP: Serine Protease; FDA: U.S. Food and Drug Administration;
hACE2: Human Angiotensin-converting Enzyme 2
The
authors declare that they have no discernible competing financial interests or
personal connections that could be interpreted as influencing the conclusions
made in this work.
13. Funding
This
research received no specific grant from any funding agency in the public,
commercial or not-for-profit sectors.
14. Acknowledgment
Infrastructure facilities provided by the Department of
Biochemistry, Jawaharlal Nehru Medical College, under the DST (FIST &
PURSE) program are gratefully acknowledged.
15. CRediT
authorship contribution statement
Mohd Mustafa:
Conceptualization, Data Curation, Investigation, Writing - Original Draft
Preparation; Kashif Abbas:
Methodology, Data Curation, Formal Analysis; Waleem Ahmad: Supervision, Data Curation, Investigation; Rizwan Ahmad: Data Curation,
Investigation; Sidra Islam:
Methodology, Writing-Review & Editing; Irfan Qadir Tantry: Funding Acquisition, Resources; Moinuddin: Supervision,
Conceptualization, Writing-Review & Editing; Md. Imtaiyaz Hassan: Funding Acquisition, Supervision,
Visualization; Mudassir Alam:
Data Curation, Formal Analysis; Nazura
Usmani: Funding Acquisition, Supervision, Formal Analysis; Safia Habib: Conceptualization, Funding
Acquisition, Resources, Supervision, Writing-Review & Editing.
16. References
12. Gralinski LE, Menachery VD. Return of the
Coronavirus: 2019-nCoV. Viruses, 2020;12: 135.
49. Sit TH, Brackman
CJ, Ip SM, et al. Canine SARS-CoV-2 infection. Nature, 2020;586: 776.
53. Liu
DX, Liang JQ, Fung TS. Human coronavirus-229E, -OC43, -NL63 and -HKU1
(Coronaviridae). Encyclopedia of Virology, 2021;428.